Calculating Genetic Distances between Protein Sequences
Procedure to work simulator
- Consider three random protein sequences: Protein Sequence 1, Protein Sequence 2, Protein Sequence 3.

.fasta format aminoacid sequences were aligned using multiple sequence alignment with sequinR package. The dist.alignment() function takes a multiple alignment as input and calculates calculates the genetic distance between each pair of proteins in the multiple alignment.
- Click on run button to execute simulator.
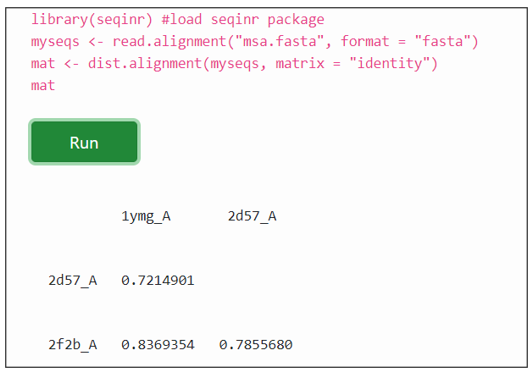
The numerical values in matrix of the output indicates genetic distance between each pair of proteins in the multiple alignment. The larger the genetic distance between two sequences, the more amino acid changes or indels that have occurred since they shared a common ancestor, and the longer ago their common ancestor probably lived.
DIY
Follow ( https://vlab.amrita.edu/index.php?sub=3&brch=311&sim=1835&cnt=2) to install R in personal computer.
Install the SeqinR package.
Import “seqinr” library to R workspace
Create a function() for retrieving multiple sequence from database
Function 1
create a list to store sequences connect to the database for each element in the array query the sequence and assign to a variable append the data into a list end close the connection return list end of fuction 1
Create a vector of sequences
Retrieve the sequences from the database
Write the sequences into a file
Align the sequence and assign to a variable in phylip format