
Phylogenetic Tree Reconstruction Using Fitch-Margoliash Algorithm
1. Upload Data File: Click "Choose file" to upload your input file (.txt/FASTA format)
2. Click "Sample Text file" to obtain a sample text file format or Click "Sample FASTA" to obtain a sample FASTA file format
3. Click to 'Run Algorithm' to get the result.
4. Click to 'Next Step' to do clustering.
6. Reset to start over
Step 1: Compute the Pairwise Distance Matrix
Step 2: Identify the Closest Pair of Taxa
Step 3: Create a new internal node that represents the common ancestor of the closest taxa.
▪Assign branch lengths to the new node using the weighted least-squares method.
▪The branch length from taxa to the new node is computed as: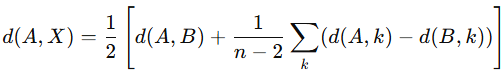 Where 𝑋 is the new node, and 𝑛 is the total number of taxa.
Where 𝑋 is the new node, and 𝑛 is the total number of taxa.
▪Assign branch lengths to the new node using the weighted least-squares method.
▪The branch length from taxa to the new node is computed as:
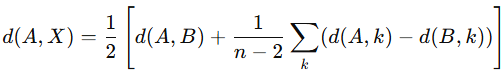 Where 𝑋 is the new node, and 𝑛 is the total number of taxa.
Where 𝑋 is the new node, and 𝑛 is the total number of taxa.
Step 4: Update the Distance Matrix
▪Compute distances from X to all other taxa using the formula:
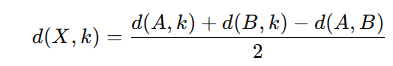
Step 5: Repeat Steps 2-4 Until Only One Node Remains
Step 6: Construct the Phylogenetic Tree